por
Lauren Dubinsky, Senior Reporter | February 23, 2018
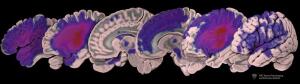
This image depicts the distribution
of lesions in the dataset.
The redder the color, the more patients
had lesions in that region.
The largest open-source data set of brain MR exams of stroke patients is now available for download.
To date, 33 research groups around the world have downloaded the Anatomical Tracings of Lesions After Stroke (ATLAS) data set to develop and test algorithms that automatically process MR images of stroke patients.
"The fact that so many researchers from around the world were willing to share their stroke MR data with us to create a public resource reflects a recent and positive change in how researchers view scientific collaboration," Sook-Lei Liew, first author of the study and assistant professor at the Mark and Mary Stevens Neuroimaging and Informatics Institute, told HCB News.



Ad Statistics
Times Displayed: 132490
Times Visited: 7530 MIT labs, experts in Multi-Vendor component level repair of: MRI Coils, RF amplifiers, Gradient Amplifiers Contrast Media Injectors. System repairs, sub-assembly repairs, component level repairs, refurbish/calibrate. info@mitlabsusa.com/+1 (305) 470-8013
Data sharing has historically been hindered by competition between research groups and limited resources to collect data. However, emerging artificial intelligence computation methods require large data sets to train and test the algorithms.
"Funding agencies and scientific communities are now recognizing the value of team science and data sharing for furthering scientific advancements," said Liew. "This means that researchers are becoming more open with data sharing, and realizing that sharing data is the best way to generate these powerful large data sets."
Liew and her team plan to use the algorithms to perform a meta-analysis of thousands of stroke MRs, to better understand how lesions affect recovery. That information will help scientists identify biological markers that can predict which patients will respond to different rehabilitation therapies and personalized treatment plans.
Scientists currently have to manually draw boundaries around lesions, which would be an impossible task to perform on thousands of images. The algorithms under development will be able to automate that process.
Liew and her team are currently testing all existing algorithms that attempt to automate the process in order to determine which results in the greatest accuracy. They found that a toolbox called Lesion Identification with Neighborhood Data Analysis (LINDA) performed the best.
"However, even the best segmentation algorithms still fail on a number of lesion cases, so we strongly emphasize the use of visual inspection of all segmentation outputs, followed by manual quality control," said Liew. "Using an automated algorithm, followed by visual quality control and manual editing still saves considerable time over doing everything manually."
Back to HCB News

